Artrose onderzoek
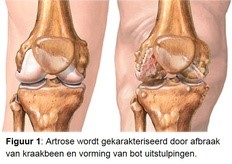
Artrose (figuur 1) is de meest voorkomende chronische ziekte in ons land. Maar liefst 1.2 miljoen mensen hebben artrose en kampen dagelijks met de pijn en bewegingsbeperkingen die artrose met zich meebrengt. Naast een gewrichtsvervangende operatie in het eindstadium van de ziekte, is er geen werkend medicijn. Patiënten zijn dan ook jarenlang afhankelijk van fysiotherapie en palliatieve zorg. Risicofactoren van artrose zijn leeftijd, overgewicht, mechanische overbelasting en erfelijke aanleg.
Wat is het probleem?
Een belangrijk probleem die de ontwikkeling van medicijnen remt is dat de farmaceutische industrie tot nu toe heeft gezocht naar een medicijn voor de gemiddelde artrosepatiënt. Maar die bestaat niet. Artrose ontstaat door verschillende oorzaken (bijvoorbeeld erfelijke aanleg, overbelasting, overgewicht) en de ziekte uit zich op verschillende wijzen. Welke patiënt welke vorm van artrose heeft is nog niet goed onderzocht en is lastig te herkennen. Zelfs niet met behulp van moderne apparatuur zoals de MRI. De verschillende ziekteprocessen ontstaan omdat de cellen in het gewricht verschillend reageren op schade aan het weefsel.
De RAAK studie
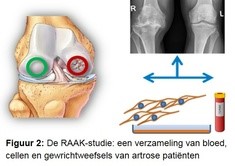
In het onderzoek maken we gebruik van de RAAK-studie (figuur 2). In deze studie hebben we bloed en gewrichtweefsels van artrose patiënten verzameld tijdens een gewrichtsvervangende operatie. De weefsels moeten namelijk worden verwijderd om het kunstgewricht te kunnen plaatsen. Omdat de gewrichtweefsels (bot, kraakbeen, slijmvlieslaag) direct na de operatie worden opgehaald kunnen we ook levende cellen uit de weefsels halen.
Het onderzoek
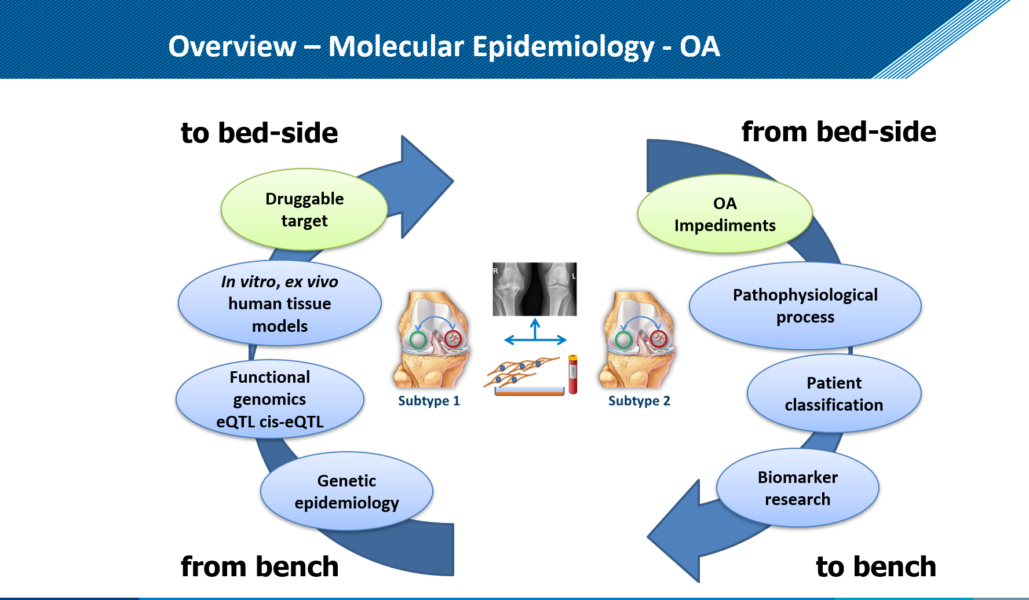
Diversiteit in kaart brengen Wij willen de diversiteit van de moleculaire paden, ofwel de ziekteprocessen, die tot artrose leiden in kaart brengen en daarmee verschillende patiëntgroepen van elkaar onderscheiden. We bestuderen daarom in de gewrichtweefsels, de erfelijke opmaak, de activiteit van eigenschappen (genen) en het mechanisme van cellen om de activiteit van deze eigenschappen te controleren.
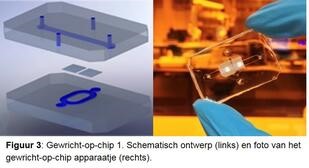
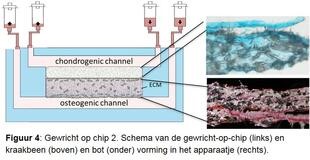
Artrose ziekteproces nauwkeurig onderzoeken. Om de verschillende ziekteprocessen nauwkeurig te bestuderen ontwikkelen we samen met de TU Eindhoven een gewricht-op-een-chip (figuren 3 en 4). Dit is een minuscule apparaatje die twee met elkaar verbonden ruimtes heeft. De ene wordt gevuld met kraakbeencellen, de andere met botcellen van de RAAK studie en met behulp van groeifactoren wordt nieuw kraakbeen en bot weefsel in het apparaatje aangemaakt. Doordat we de cellen blootstellen aan bijvoorbeeld mechanische belasting, door met piepkleine hamertjes op de cellen te slaan, kunnen we het artrose proces nabootsen. We veranderen ook de genetische opmaak van de cellen, om patiënt specifieke oorzaken te onderzoeken. Door nauwkeurig vast te stellen waar het mis gaat tijdens artrose in verschillende patiënten, krijgen we ook aanknopingspunten voor de ontwikkeling van (nieuwe) medicijnen. Dit noemen we ook behandeling op maat.
Signaalmoleculen om ziekteproces waar te nemen
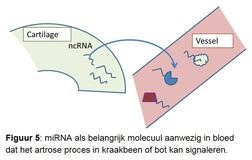
Er is geen adequate diagnostiek voorhanden die het precieze artroseproces in de gewrichtsweefsels kan waarnemen en volgen in de tijd. Dit levert grote problemen op bij het uittesten van nieuwe medicijnen. Het is namelijk niet mogelijk om patiënten te selecteren die, op basis van het aanwezige ziekteproces, waarschijnlijk het meeste gebaat zijn bij het specifieke aangrijpingspunt van het ontwikkelde medicijn. Het onderzoek is er daarom op gericht om een veelbelovend nieuw moleculair signaalmolecuul te gebruiken. Het gaat hier om zogenaamde micro-RNAs. Micro-RNAs zijn kleine stukjes genetisch materiaal die zorgen voor de regulatie en controle van activiteit van genen en daarmee van specifieke processen in weefsels (figuur 5). Uniek is dat micro-RNAs via de bloedsomloop ook zorgen voor het uitwisselen van berichten over de toestand van de weefsels in het lichaam. Dit is het moment waarop we de micro-RNAs en hun boodschap kunnen onderscheppen en gebruiken als biomarker.
Meest Recente Publicaties Artrose
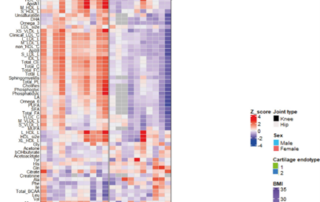
New publication shows blood biomarkers for senescence endotypes in osteoarthritis patients
When cells reach a permanent growth arrest (Senescence) and resist death, this can result in numerous diseases, such as osteoarthritis. Here we show the presence of two different types of senescence in the blood metabolome of osteoarthritis patients. These blood metabolome patterns could function [...]
A human in vitro 3D neo-cartilage model to explore the response of OA risk genes to hyper-physiological mechanical stress
A human in vitro 3D neo-cartilage model to explore the response of OA risk genes to hyper-physiological mechanical stress | Osteoarthritis and Cartilage Open| 4(1) (2022) 100231 | doi: 10.1016/j.ocarto.2021.100231 R.G.M. Timmermans, N.G.C. Bloks, M. Tuerlings, M. van Hoolwerff, R.G.H.H. Nelissen, R.J.P. van der [...]
A molecular map of long non-coding RNA expression, isoform switching and alternative splicing in osteoarthritis
A molecular map of long non-coding RNA expression, isoform switching and alternative splicing in osteoarthritis | Hum Mol Genet | 2022 Jan 28:ddac017 | doi: 10.1093/hmg/ddac017. Katsoula G, Steinberg J, Tuerlings M, de Almeida RC, Southam L, Swift D, Meulenbelt I, Wilkinson JM, Zeggini [...]
ARTROSE TEAM